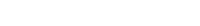Hi Philippe,
Thank you very much for cluing me into that flag. That in fact did get me over that hurdle. I was intently focused on the problem somewhere with my domain (block sizes, max blocks and processor types) that I failed to review the 'setup' namelist as a potential culprit.
The result of your help is an attached graphic of sea ice concentration modelled using CICE6 forced from re-gridded ERA5 to 1/10-degree tripole B-grid with an hourly time-step -- this daily mean contains no ocean forcing and started with no initial conditions -- hence the results are not real representation of global sea ice concentration for this historical date.
 You or someone else might care to add another comment on CICE6 standalone model processing duration?
You or someone else might care to add another comment on CICE6 standalone model processing duration?
The attached graphic was a result of roughly one hour of processing time -- i.e. one week's worth of model output in one hour of processing. I do not have a good gauge on whether this is a reasonable value? Note, I'm currently configured using OpenMP intel compilers on 48 processors and 2990GB of memory -- the large memory is for 1.2TB atmosphere forcing file. Anyhow, there is a lot to unpack in the details of the documentation between
3.1.2 and
3.1.4.3 that have to do with performance of the model as well as optimisation of my particular area of interest -- Antarctic landfast ice. I realise I'll also should give a more thorough consideration
here (3.2.3.1), and engage/consult with the administrators of the super computer I'm using.
Regardless, thank you again cluing me into to essentially a very easy 'flip-of-the-switch' fix!
--------------------------------------------------------------------------------------------------------
Another question, along the same thread of 'sandboxing' (at least my definition of that term), the 'segfault' crash that I
posted previously, do you have any thoughts on the nature of that crash?
I'm now attempting to load in the ocean forcing file via
ocean, which, is in fact, a derived (and re-gridded) BlueLink ReANlysis (BRAN) dataset that mimics the structure required by
subroutine ocn_data_ncar_init
in file
ice_forcing.F90
Interestingly the crash is happening here:
ocean mixed layer forcing data file =
/scratch/jk72/da1339/cice-dirs/input/AFIM/forcing/0p1/daily/bran_ocn_frcg_cice6
_2005.nc
tracer index depend type has_dependents
hi 1 0 1 T
hs 2 0 1 T
nt_Tsfc 3 0 1 F
nt_qice 4 1 2 F
nt_qsno 11 2 2 F
nt_sice 12 1 2 F
nt_iage 19 1 2 F
nt_FY 20 0 1 F
nt_alvl 21 0 1 T
nt_vlvl 22 1 2 F
nt_apnd 23 21 2 T
nt_hpnd 24 23 3 F
nt_ipnd 25 23 3 F
nt_fbri 26 0 1 F
nt_smice 26 0 1 F
nt_smliq 26 0 1 F
nt_rhos 26 0 1 F
nt_rsnw 26 0 1 F
nt_fsd 26 0 1 F
nt_isosno 26 0 1 F
nt_isoice 26 0 1 F
nt_bgc_S 26 0 1 F
Find indices of diagnostic points
found point 1
lat lon TLAT TLON i j block task
-65.0 0.0 -65.0 0.0 27 89 6 9
found point 2
lat lon TLAT TLON i j block task
-65.0 -179.0 -65.0 -179.0 36 89 14 3
(calc_timesteps) modified npt from 168 1 with dt= 3600.00
(calc_timesteps) to 168 1 with dt= 3600.00
(calc_timesteps) start time is 2005-01-01:00000
(calc_timesteps) end time is 2005-01-08:00000
Initial forcing data year = 2005
Final forcing data year = 2005
Atmospheric data files:
/scratch/jk72/da1339/cice-dirs/input/AFIM/forcing/0p1/JRA55/8XDAILY/JRA55_gx3_0
3hr_forcing_2005.nc
Set current forcing data year = 2005
(JRA55_data) reading forcing file 1st ts =
/scratch/jk72/da1339/cice-dirs/input/AFIM/forcing/0p1/JRA55/8XDAILY/JRA55_gx3_0
3hr_forcing_2005.nc
Finished writing ./history/iceh_ic.2005-01-01-03600.nc
[gadi-mmem-clx-0002:727506:0:727506] Caught signal 11 (Segmentation fault: Sent by the kernel at address (nil))
[gadi-mmem-clx-0002:727472:0:728081] Caught signal 11 (Segmentation fault: Sent by the kernel at address (nil))
...skipping 69 lines
cice 00000000008A3585 ice_transport_rem 576 ice_transport_remap.F90
cice 000000000088F64E ice_transport_dri 545 ice_transport_driver.F90
cice 00000000004142AC cice_runmod_mp_ci 297 CICE_RunMod.F90
cice 000000000040E806 MAIN__ 49 CICE.F90
cice 000000000040E79D Unknown Unknown Unknown
libc-2.28.so 000014D062535CF3 __libc_start_main Unknown Unknown
cice 000000000040E6BE Unknown Unknown Unknown
forrtl: error (78): process killed (SIGTERM)